| Journal of Neurology Research, ISSN 1923-2845 print, 1923-2853 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Neurol Res and Elmer Press Inc |
| Journal website https://www.neurores.org |
Original Article
Volume 14, Number 2, December 2024, pages 49-58
Assessing Internal Reproducibility Within a Parkinson’s Disease Cohort by Leveraging an Independent Larger Dataset
Kristen Watkinsa, b, g, Julia Greenberga, g, Kelly Astudilloa, c, Charalambos Argyroub, d, Wen-Yu Leee, John F. Craryf, Steven J. Fruchta, c, Towfique Rajb, d, Giulietta Maria Riboldia, c, h
aDepartment of Neurology, New York University Langone Health, New York, NY, USA
bNash Family Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA
cThe Marlene and Paolo Fresco Institute for Parkinson’s Disease and Movement Disorders, New York University Langone Health, New York, NY, USA
dRonald M. Loeb Center for Alzheimer’s Disease, Icahn School of Medicine at Mount Sinai, New York, NY, USA
eDepartment of Population Health, New York University Langone Health, New York, NY, USA
fDepartments of Pathology, Neuroscience, and Artificial Intelligence and Human Health, Neuropathology Brain Bank and Research Core, Ronald M. Loeb Center for Alzheimer’s Disease, Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA
gThese authors contributed equally to this work.
hCorresponding Author: Giulietta Maria Riboldi, The Marlene and Paolo Fresco Institute for Parkinson’s and Movement Disorders, New York University Langone Health, New York, NY 10017, USA
Manuscript submitted October 18, 2023, accepted August 22, 2024, published online November 15, 2024
Short title: Clinical Clusters in Parkinson’s Disease
doi: https://doi.org/10.14740/jnr761
| Abstract | ▴Top |
Background: Parkinson’s disease (PD) is a complex and heterogeneous disorder that is likely composed of several phenotypic subgroups with distinct clinical features and patterns of disease progression. Cluster analysis, which categorizes subjects into groups of “maximal similarity”, is a valuable statistical tool for characterizing phenotypic variability in clinical cohorts and for correlating phenotypes with specific biomarkers. However, data collection methods often differ between clinical and research settings, limiting the ability to obtain statistically significant results from smaller or less characterized cohorts and to compare results across studies. Establishing reproducibility of clinical cluster analysis across different studies/centers would allow generalizability across studies. The goal of this study was to leverage cluster analysis of clinical traits to establish reproducibility of clinical phenotypes in a cohort of patients with PD at local centers (Discovery cohort) and the large PD bioregistry Parkinson’s Progression Markers Initiative (PPMI cohort).
Methods: Nonhierarchical k-means clustering by phenotype of subjects in the Discovery (n = 179) and PPMI (n = 368) cohorts was performed via principal component analysis (cohort-based clusters). Eigenvectors of clustering in the PPMI cohort were identified and utilized to re-cluster the Discovery cohort (PPMI-based clusters). Overlap in cluster membership between cohort-based clusters and PPMI-based clusters of the Discovery cohort was assessed.
Results: Clustering of subjects revealed two clusters in the Discovery cohort and three clusters in the PPMI cohort. The first four principal components for clustering of the PPMI cohort, accounting for 43% of the variability, were driven by depression, anxiety, age at symptom onset, gender, and a tremor-dominant phenotype. After re-clustering the Discovery cohort based on these traits, 89% of subjects remained in their original cluster (κ = 0.776, P < 0.01).
Conclusions: We successfully leveraged cluster analysis of clinical traits in PD patients from the larger and standardized PPMI cohort to validate reproducibility of clustering in our smaller Discovery cohort. We propose a combination of nonhierarchical cluster analysis and testing of generalizability with re-clustering to establish clustering reproducibility. This method can be adapted for use in a wide range of clinical scenarios, allowing for analysis of cohorts that are less extensively characterized or those with low intrinsic power secondary to low sample size.
Keywords: Parkinson’s disease; Cluster analysis; Cross-validation; Phenotype; PPMI
| Introduction | ▴Top |
Parkinson’s disease (PD) is a heterogeneous disorder, likely composed of several phenotypic subgroups with distinct clinical features and patterns of disease progression [1, 2]. Delineating these phenotypes has important implications for understanding disease pathophysiology, prognostication, and future therapeutics.
Cluster analysis, which categorizes subjects into groups of “maximal similarity”, serves as a valuable statistical tool for characterizing phenotypic variability in PD cohorts and for correlating phenotypes with specific biomarkers. Many variations on this method have been applied to different cohorts of PD patients in an effort to discover distinct phenotypic subgroups (i.e., motor dominant vs. nonmotor dominant; tremor dominant (TD) vs. rigid akinetic) [1, 2]. However, data collection methods often differ between clinical and research settings, limiting the ability to use cluster analysis to obtain significant results from smaller or less-well characterized cohorts, as well as to compare findings across studies. For example, a recent systematic review of 20 PD clustering studies found that, across studies, there was a wide variety in the variables included, statistical methods used, numbers of clusters found (two, three, four, or five), and characteristics of the resulting clusters [3]. This limitation was further highlighted by Mestre et al [4], who looked at published subtype classification systems produced by data-driven clustering of eight separate PD cohorts, and was only able to reproduce the classifications of one of those eight studies in a well-established reference cohort (the Longitudinal and Biomarker Study in Parkinson’s Disease (LABS-PD)). The researchers raised the concern that there were gaps in validity of currently defined subtypes and concluded that implementing external validation should be standard practice when reporting new subtype classification systems [4]. Establishing reproducibility of clinical cluster analysis across studies would greatly expand the generalizability, and thus clinical utility, of this tool for characterizing phenotypic variants in PD.
The Parkinson’s Progression Markers Initiative (PPMI) comprises a large PD observational study and biorepository, with comprehensive clinical, genetic, imaging, and blood biomarker data from approximately 1,500 subjects. Large, multicenter, longitudinal PD biorepositories can be leveraged for cluster analysis to establish reproducibility in smaller, previously unvalidated cohorts, as described above [1, 4, 5]. The present study compares cluster analysis of the PPMI cohort with the PD cohort at our centers to establish the reproducibility of the clustering solution of our cohort.
| Materials and Methods | ▴Top |
Data collection
The Discovery cohort consisted of patients with PD at the Mount Sinai Bendheim Parkinson and Movement Disorder Center (MSMD) and The Marlene and Paolo Fresco Institute for Parkinson’s and Movement Disorders at NYU Langone (NYU). MSMD and NYU cohort data were collected via chart review and interview, respectively (Supplementary Material 1, www.neurores.org). The following demographic data were collected: gender, current age, age of onset, disease duration, and family history of PD. Current age was excluded from analyses due to tight positive correlation with age at symptom onset. The following binary clinical data were collected: presence of freezing of gait (FoG), dyskinesias, motor fluctuations, rapid eye movement (REM)-sleep behavior disorder (RBD), autonomic symptoms (orthostasis, urinary symptoms, constipation), neuropsychiatric symptoms (depression, anxiety, dementia, hallucinations), hyposmia, inflammatory comorbidities (e.g., asthma, rheumatoid arthritis), use of PD-related medications (i.e., dopaminergic medications, monoamine oxidase inhibitors (MAOi), catechol-O-methyltransferase (COMT) inhibitors) and anti-inflammatory medications (aspirin), and motor phenotype as assessed clinically (i.e., TD phenotype vs. postural instability and gait disorder (PIGD)). Characterization of binary data was based on clinical judgment. Genetic variances in the glucocerebrosidase (GBA) and leucine-rich repeat kinase 2 (LRRK2) genes were collected as well. Nonbinary data collected included modified Hoehn and Yahr scale (H&Y), University of Pennsylvania Smell Identification Test (UPSIT), Unified Parkinson’s Disease Rating Scale Part III (UPDRS III), and Montreal Cognitive Assessment (MoCA) [6]. Analysis of the Discovery cohort was limited to subjects with idiopathic PD and age of onset greater or equal to 40 years old to exclude subjects with early onset PD, who have distinct phenotypic presentations and disease-related pathological mechanisms [7]. Subjects with over 50% data missingness and traits with over 25% data missingness were excluded. Missing values for the remaining traits and subjects were then imputed using the MICE (v3.8.0) and VIM (v5.1.1) packages, and disease duration was regressed out [8, 9]. In particular, since our dataset included continuous and ordinal variables, we coded logistic or ordinal regression (based on the type of data) to obtain residual. Then, standardized residual was calculated for each variable. Rstudio v2024.04.2+764 (R version 4.4.1) was used for all analyses [10].
For the PPMI cohort, PPMI data were downloaded from the Laboratory of Neuroimaging (LONI) database (2018 data cut). PPMI is a multicenter longitudinal observational study that consists of healthy controls and subjects with PD [11]. Inclusion criteria for the PD cohort included: diagnosis of PD within 2 years of enrollment and naivety to dopaminergic drugs at enrollment. All demographic, motor, and nonmotor data used in these analyses were downloaded from the first available datapoint (screening and baseline visit). The following demographic data were considered in the analyses: current age, age at diagnosis, sex, and family history of PD. The following rating scales assessing for the motor symptoms of PD were considered in the analyses: H&Y, and Movement Disorder Society - Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) Part III. The following rating scales for PD-associated nonmotor symptoms were considered in the analyses: Geriatric Depression Scale (GDS) Short Form, State-Trait Anxiety Inventory (STAI), Scales for Outcome in Parkinson’s Disease - Autonomic (SCOPA-AUT), REM Sleep Disorder Questionnaire (RBD-SQ), Epworth Sleepiness Scale (ESS), Questionnaire for Impulsive-Compulsive Disorders in Parkinson’s Disease (QUIP), UPSIT, and MoCA. Our analyses were limited to all subjects in the PPMI PD cohort with age of onset greater or equal to 40 years old [7]. Any subjects that had missingness for any of the included demographic factors or scales were excluded. All continuous and discrete data as well as the H&Y scale were scaled using z-scores [1, 12, 13]. Disease duration was not controlled for as all subjects were enrolled within 2 years of symptom onset. TD and PIGD subscores were calculated using UPDRS (off state) data from the baseline visit as previously described [14].
Cluster analysis of discovery cohort
Principal component analysis (PCA) was applied to the Discovery cohort data using the stats package prcomp function (stats v4.4.1) [10]. The results of this analysis were entered into a nonhierarchical k-means clustering analysis, specifying the optimal number of clusters, to identify subjects’ cluster membership. The optimal number of clusters was determined using the NbClust package (v3.0.1), which uses 23 indices to propose the best clustering scheme based on majority rule. The function necessitates manual input of minimum and maximum number of clusters the function could suggest, which in these analyses were two through seven inclusive, as well as method type, of which “k-means” was selected. If the package’s first best recommendation was deemed not clinically relevant, as determined by degree of cluster overlap and number of subjects per cluster, then the second-best recommendation was selected [15].
To keep data type consistent in the Discovery cohort data set, in which most data were binary, non-binary data were converted into categorical data. Disease duration was divided into < 5 years, 5 - 9 years, and ≥ 10 years; age at onset was divided into < 50, 50 - 59, 60 - 69, ≥ 70; H&Y was divided into < 3 or ≥ 3.
Chi-square and unpaired t-tests were used to compare categorical and continuous variables, respectively, with Bonferroni correction and a significance cutoff of adjusted P < 0.05.
Cluster analysis of PPMI cohort
As performed with Discovery cohort, PCA followed by k-means clustering analysis was applied to the standardized PPMI cohort data, using NbClust to determine the optimal number of clusters. Chi-square and analysis of variance (ANOVA) were used to compare categorical and continuous variables, respectively, with Bonferroni correction and a significance cutoff of adjusted P < 0.05.
To determine the number of principal components (PC) to retain for future analyses, the eigenvalues of (or portion of variance explained by) each PC were visualized using a scree plot. Using the “elbow” method, only data from those PCs up to the inflection point were retained. Rotated loadings of the retained PCs were used to identify variables that contributed significantly to the eigenvectors. For each PC, the two traits with the greatest factor loading (absolute value) were selected.
Determining reproducibility
The Discovery cohort data were then re-entered into PCA, using only the variables that contributed most to clustering results of the PPMI cohort based on eigenvectors’ loadings, as reported above. K-means clustering analysis was applied; NbClust was used to determine the optimal number of clusters. Overlap in cluster membership between the original Discovery cohort clustering results (cohort-based clustering) and new Discovery cohort clustering results (clustering agreement) was assessed by calculating the percent of subjects, whose cluster membership remained stable or changed. Cohen’s kappa statistic value (κ) was then calculated, comparing clusters derived from cohort-based clustering and validation-based clustering, with a nominal significant cutoff of P < 0.05 [16, 17].
Genetic analysis
Samples were screened through targeted genotyping for the G2019S variant of the LRRK2 gene, and for the following variants for the GBA gene: IVS2+1, 84GG, E326K, T369M, N370S, V394L, D409G, L444P, A456P, R496H, RecNcil. The variants were selected based on frequency in the PD population and among subjects with Ashkenazi Jewish ancestry. Analyses were performed at Dr. William Nichols’ Laboratory at the Cincinnati Children’s Hospital.
Institutional Review Board (IRB) approval and ethical compliance statement
All study procedures were reviewed and approved by New York University’s IRB and the Mount Sinai Hospital IRB. This study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
| Results | ▴Top |
Cluster analysis of the Discovery cohort resulted in two clusters differentiated by motor and nonmotor symptoms
The Discovery cohort included 198 subjects, including 66 from the NYU clinic and 132 from the MSMD clinic. Nineteen subjects were excluded due to age of onset under 40. Final sample size was 179 subjects, of which 63% of subjects were male, with mean age 68.7 ± 8.5 years and mean disease duration 8.8 ± 5.1 years, including 28 carriers of disease-associated variants of GBA (18 subjects with N370S variant, one 84GG, one 84GG/T369M, three E326K, one L444P/A456P/RecNcil, two R496H, one RecNcil, one T369M, one V394L) and 12 carriers of disease-associated variants of LRRK2 (G2019S) (Table 1). Four subjects carried both the LRRK2 G2019S variant and the GBA N370S variant - two were included in the GBA-positive group and two in the LRRK2-positive group (Table 1).
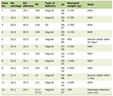 Click to view | Table 1. Demographic Characteristics for PPMI and Discovery Cohorts at Baseline |
As data from the MSMD subset of the Discovery cohort were collected via retrospective chart review, not all data points and written scales were available for these subjects. The following variables were excluded from analysis due to meeting the exclusion criteria of greater than 25% missingness (percent missingness noted in parenthesis): UPSIT (80% missingness), subjective hyposmia (32%), UPDRS III (68%), UPDRS total (66%), and MoCA (66%) (Supplementary Material 2, www.neurores.org). Analysis to determine the optimal number of clusters for the Discovery cohort resulted in six indices recommending a two-cluster solution (Fig. 1a). Nine indices proposed seven as the best number of clusters. However, because of the small sample size of our cohort, the seven-cluster solution would not be able to identify clinically significant groups. Therefore, the two-cluster solution was chosen for the downstream analysis. Cluster 2 (n = 87) was characterized by higher rates of anxiety (84% vs. 7.6%, P ≤ 0.001) and depression (82% vs. 13%, P ≤ 0.001) and a trend towards older age of onset (61.4 ± 9.0 vs. 58.5 ± 9.6, P = 0.735). The difference between the other traits was not statistically significant, but there was an over representation of TD phenotype in cluster 1 (n = 92) and of dysautonomia, PIGD subtype, and nonmotor symptoms in cluster 2 (Fig. 1b, Table 2, Supplementary Material 3, www.neurores.org) [13].
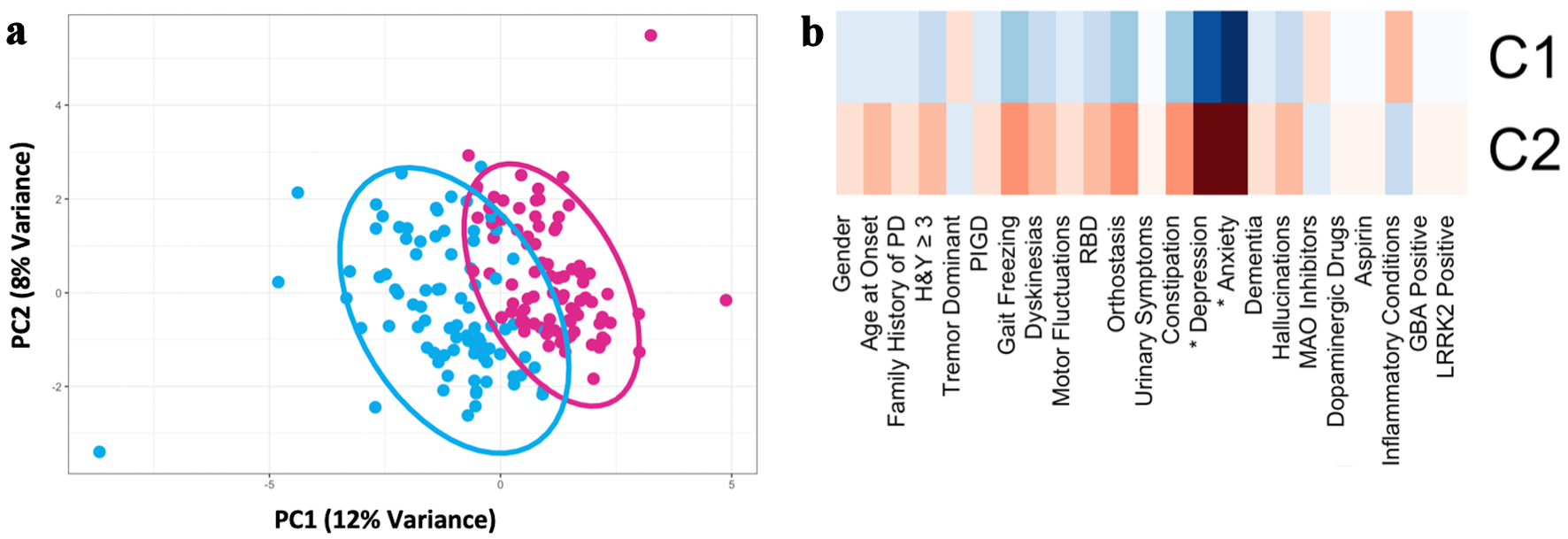 Click for large image | Figure 1. Principal component analysis of the discovery cohort (n = 179) based on demographic, motor, and nonmotor characteristics demonstrated two PD subtypes (after imputing missing data and controlling for disease duration). (a) Scatter plot depicting separation of subjects across the first two principal components. (b) Heatmap of clinical characteristics for each cluster identified. The greater red hue indicates greater prevalence, greater blue hue indicates lower prevalence. Traits with a statistically significant difference between clusters, defined as Bonferroni corrected P < 0.05, are indicated with an asterix (*). PD: Parkinson’s disease; RBD: REM-sleep behavior disorder; MAO: monoamine oxidase; PIGD: postural instability and gait disorder; GBA: glucocerebrosidase; LRRK2: leucine-rich repeat kinase 2; H&Y: Hoehn and Yahr scale; PC: principal component. |
 Click to view | Table 2. Comparison of Discovery Cohort Clusters |
Cluster analysis of the PPMI cohort resulted in three clusters differentiated by age of onset, motor symptoms, and psychiatric features
To compare the cluster analysis results of our discovery cohort with a large and standardized database, the same cluster analysis methods were repeated with the PPMI cohort. The PPMI cohort included 433 subjects. Fifty-seven (13%) of subjects were excluded due to data missingness. The most common missing clinical data were family history (23 subjects). RBDSQ (16 subjects), and STAI (7 subjects); additionally, age and gender were not inputted for 12 and seven subjects respectively (Supplementary Material 4, www.neurores.org). Fourteen subjects were excluded due to age of onset under 40. Final PPMI cohort sample size was 368 newly diagnosed, medication-naive subjects with PD, of which 66% were male, with mean age at baseline visit 61.9 ± 8.8 years and mean disease duration at baseline 6.5 ± 6.6 months (range 0 - 36 months) (Table 1).
Analysis of the optimal number of clusters for the PPMI cohort resulted in 12 indices recommending a three-cluster solution (Fig. 2a). Cluster 1 (n = 71) was characterized by more severe anxiety (P < 0.01), depression (P < 0.01), impulse control disorder (P < 0.01), and orthostasis (P < 0.01). Cluster 2 (n = 142) was characterized by a less severe phenotype considering motor and nonmotor symptoms. Cluster 3 (n = 155) was characterized by older age of onset (P < 0.01), greater impairment on MDS-UPDRS part 3 (P < 0.01), and higher H&Y (P < 0.01) (Fig. 2b, Table 3) [13].
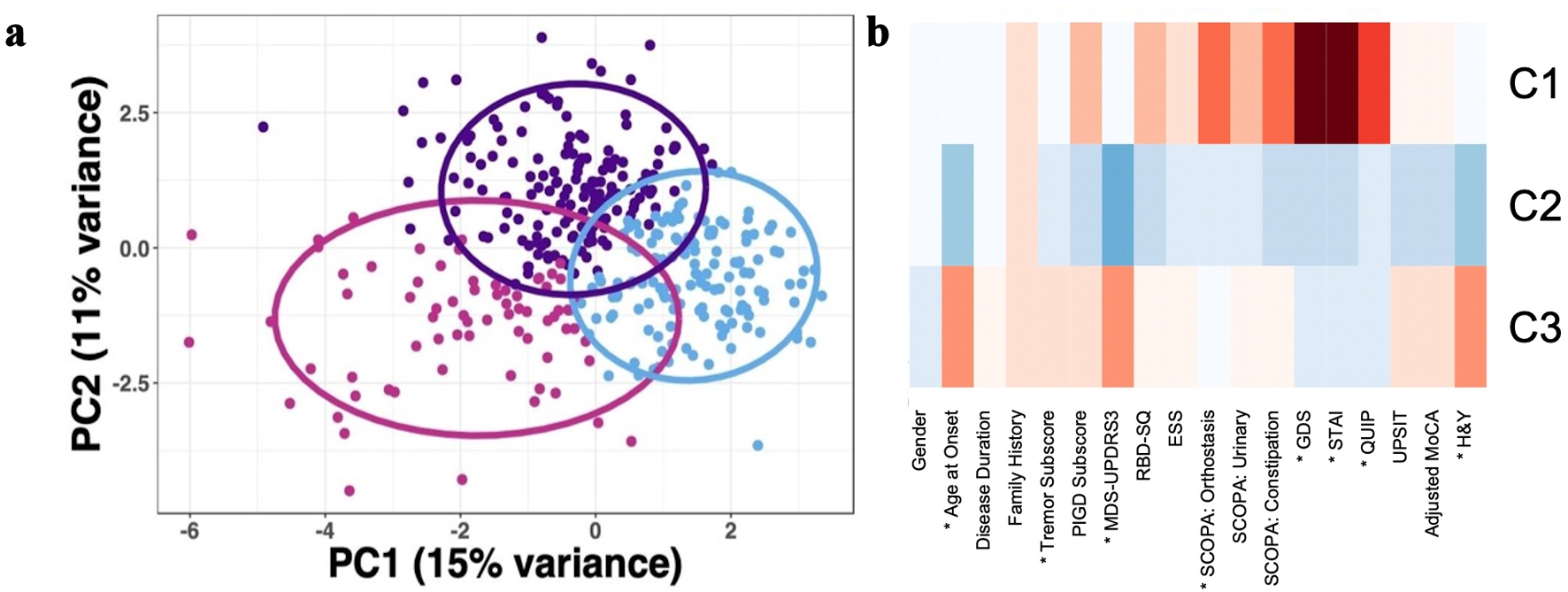 Click for large image | Figure 2. Principal component analysis of the PPMI cohort (n = 368) based on demographic, motor, and nonmotor characteristics demonstrated three PD subtypes. (a) Scatter plot depicting separation of subjects across the first two principal components. (b) Heatmap of clinical characteristics for each cluster identified, with data scaled using z-scores. The greater red hue indicates more severe impairment or greater number of years; greater blue hue indicates less severe impairment or fewer number of years. Traits with a statistically significant difference between clusters as determined via ANOVA for continuous data and Chi-square for categorical data, defined as Bonferroni corrected P < 0.05, are indicated with an asterix (*). PD: Parkinson’s disease; PPMI: Parkinson’s Progression Markers Initiative; RBD: REM-sleep behavior disorder; UPSIT: University of Pennsylvania Smell Identification Test; MDS-UPDRS: Movement Disorder Society - Unified Parkinson's Disease Rating Scale; MoCA: Montreal Cognitive Assessment; GDS: Geriatric Depression Scale; STAI: State-Trait Anxiety Inventory; SCOPA: Scales for Outcome in Parkinson’s Disease; ESS: Epworth Sleepiness Scale; QUIP: Questionnaire for Impulsive-Compulsive Disorders in Parkinson’s Disease; PC: principal component; ANOVA: analysis of variance; REM: rapid eye movement. |
 Click to view | Table 3. Comparison of Demographic and Clinical Characteristics of the Three Clusters From the PPMI Cohort |
Per the “elbow method” of interpreting PC eigenvectors, the first four PCs explained most of the variability in the data (Supplementary Materials 5, 6, www.neurores.org). The rating scales for depression (GDS) and anxiety (STAI total score) had the highest loadings for PC1. Age at onset and MDS-UPDRS part 3 score had the highest loadings for PC2. Gender and MDS-UPDRS part 3 score had the highest loadings for PC3. Tremor subscore and sleepiness score (ESS) had the highest loadings for PC4. In summary, the following traits were determined to contribute most to variability in the PPMI cohort and were utilized for downstream analysis (excluding MDS-UPDRS part 3 score and ESS score, which had no Discovery cohort corollary): depression, anxiety, age at diagnosis, gender, and tremor subscore (Supplementary Material 6, www. neurores.org).
Establishing cluster reproducibility using results from the PPMI Cohort
To establish reproducibility of the clustering results of the Discovery cohort, we repeated cluster analysis for the Discovery cohort using only the traits that drove clustering in the larger, standardized PPMI cohort (depression, anxiety, age at diagnosis, gender, and TD phenotype). Analysis to determine the optimal number of clusters for the Discovery cohort based on these five traits resulted in a two-cluster solution being recommended by six indices, and an eight-cluster solution being recommended by six indices. The two-cluster solution was deemed most relevant based on criteria described in the methods section. The two-cluster solution resulted in clusters with sample sizes of 98 and 81 subjects. Cluster 1 and cluster 2 maintained the characteristics they had after cohort-based clustering (Supplementary Materials 7, 8, www.neurores.org).
Overlap in cluster membership between the cohort-based and PPMI-based clusters of the Discovery cohort was assessed by calculating the percent of subjects whose cluster membership remained stable or changed. After re-clustering the Discovery cohort, cluster membership was reproducible in 89% of subjects (Fig. 3). Eighty-five of the 92 subjects originally in cluster 1 remained in cluster 1. Seventy-four of the 87 subjects originally in cluster 2 remained in cluster 2. Forty-two of the 179 subjects were assigned to a cluster different from their original. Cohen’s kappa statistic revealed substantial agreement (κ = 0.776, P < 0.01).
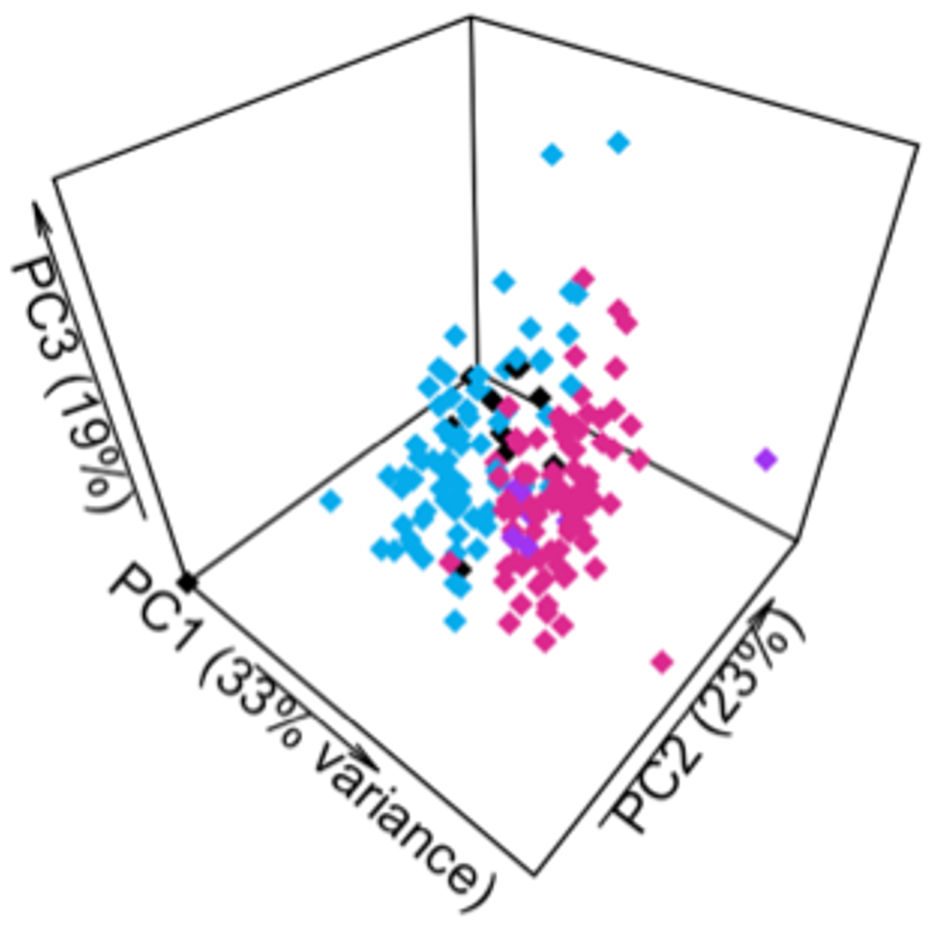 Click for large image | Figure 3. Cluster membership of subjects in Discovery cohort remained largely unchanged after re-clustering based on a limited selection of traits. Pink represents subjects assigned to cluster 1 in both analyses. Blue represents subjects assigned to cluster 2 in both analyses. Purple indicates subjects originally assigned to cluster 1 who were reassigned to cluster 2. Black indicates subjects originally assigned to cluster 2 who were reassigned to cluster 1. PC: principal component. |
| Discussion | ▴Top |
Cluster analysis of clinical cohorts in PD is a valuable tool for characterizing phenotypic variability and correlating phenotypes with specific biomarkers. From a clinical standpoint, delineating PD phenotypes helps clinicians to individualize care for PD patients, and from a translational research standpoint, it serves as a foundational step in the pathway for development of future disease-modifying therapies. However, data collection methods often differ between clinical and research settings, limiting the ability to obtain significant results from smaller or less characterized cohorts and to compare findings across studies. We successfully leveraged cluster analysis of PD subjects from one of the largest observational studies of people with PD, the PPMI study, to examine generalizability of cluster analysis results in a smaller research cohort (Discovery cohort).
Several different methods for establishing cluster reproducibility have been proposed in the literature over the years which vary depending on the aims of any particular study. Statistical models can predict the validity and reproducibility of clusters for a given data set. This approach proved particularly useful in previous works for analyzing microarrays or other large genomics datasets, in which novel clusters are discovered in the absence of pre-existing data sets that would allow for external validation [4, 5]. These models may not be ideal, however, for other scenarios, such as establishing reproducibility of cluster analyses between studies.
Similar to one such model developed by McShane et al, we used a two-step approach in our clustering analysis in which we separately conducted clustering analyses on the Discovery cohort and PPMI cohort (from the PPMI database), in order to establish a similar underlying pattern of clustering between the two groups, followed by a reproducibility analysis [18]. Our approach differed, however, in that: 1) We chose to use nonhierarchical rather than hierarchical clustering given its better relative reliability; and 2) We established reproducibility of cluster analysis for our Discovery cohort by re-clustering this cohort based on clustering analysis of the pre-existing larger, standardized PPMI cohort (i.e., an external test of reproducibility) in addition to using a statistical model like that of McShane et al or Kapp et al [18, 19]. The combination of nonhierarchical clustering and clustering agreement can represent a useful approach to establishing cluster reproducibility that can be leveraged in other cohorts.
Regarding the specific phenotypes derived from our clustering analysis, clustering of the PPMI cohort generated three groups: 1) most severe and predominantly nonmotor symptoms with trend towards a PIGD phenotype; 2) younger age of onset with overall milder symptoms (motor and nonmotor); and 3) older age of onset with predominantly motor symptoms and trend towards a TD phenotype. Previous studies have generated similar phenotypic PD subtypes using other methods of clustering analysis of PPMI data, providing further support to the reproducibility of our results [1, 2]. Fereshtehnejad et al [1], for example, identified three clusters. Their “diffuse malignant” subtype aligns with our first cluster in that both have greater impairment in GDS, STAI, QUIP, and OH; further, their study has significantly higher PIGD subscore, and ours has trend towards higher PIGD subscore, Their “mild motor-predominant” aligns with our second cluster in that both have younger age, lower UPDRS part 3, and overall mild nonmotor symptoms; to note, while in their study this cluster had the greatest proportion of TD predominant subjects, the overall tremor subscore was lowest in this cluster in both their and our study due to overall low UPDRS part 3. Lastly, their “intermediate” subtype aligns with our cluster 3 in that subjects have an older age of onset, and the scores for most nonmotor traits were between those of the other two clusters. In our study, this third cohort had the statistically significant highest UPDRS part 3, whereas in their study there was no significant difference in UPDRS part 3 between this cluster and the “mild-motor predominant” cluster [1].
Clustering of the Discovery cohort yielded two distinct phenotypes, which remained stable after re-clustering based on the PPMI cohort: 1) a less severe phenotype with milder symptoms and a trend towards TD; and 2) a more severe phenotype with prominent motor and nonmotor symptoms (trending values except for anxiety and depression that were statistically significant) (Supplementary Material 8, www.neurores.org). Promisingly, these phenotypes generally align with those obtained from clustering of the PPMI cohort, with a clear intermediate/motor predominant subtype and a severe subtype.
Although a comparison between the clusters from the PPMI cohort and the Discovery cohort would be interesting, this goes beyond the scope of this work, where the main goal was to leverage the clustering of a larger and better annotated dataset (PPMI) and assess internal reproducibility within our Discovery cohort.
There were limitations to this study. First, several variables were excluded from the initial Discovery cohort analysis due to missingness. Second, k-means clustering requires for the number of clusters to be prespecified, which can introduce bias. We minimized this bias by utilizing the NbClust package to determine the optimal number of clusters; however, it was necessary to input a range for the minimum and maximum number of clusters the function could suggest; our analyses used two to seven clusters. Lastly, limitations of our Discovery cohort included small sample size, wide variability in disease duration, lack of continuous data, and its differences compared to the PPMI cohort in traits such as average disease duration and dopaminergic medication use. These limitations, which underscore the need for leveraging larger and better characterized cohorts, likely contributed to the fact that certain cluster solutions were unstable (i.e., during the re-clustering step, only six of 30 indices recommended a two-cluster solution). Additionally, the two clusters identified were strongly differentiated by only a few traits, and that some of the traits that drove clustering in the PPMI cohort were not statistically significant drivers of clustering in the Discovery cohort.
In summary, we propose a statistical approach, consisting of nonhierarchical clustering analysis followed by assessment of clustering agreement and re-clustering to establish cluster reproducibility between our Discovery cohort and the larger, standardized database for PD (PPMI), despite differences in size and data collection methods between the two cohorts. Our results suggest that the same traits dictate cluster membership in the PPMI cohort and our Discovery cohort, adding confidence to the generalizability of future findings from the Discovery cohort to the broader PD population. The phenotypic subtypes derived from clustering analysis of the Discovery cohort align with already well-established PD phenotypes, lending further support to the reproducibility of our results. Establishing a methodology for validating reproducibility and generalizability of clustering analysis in our Discovery cohort represents the first step for further analysis in novel biomarker discovery from our cohort to be validated by the larger, standardized PPMI dataset. Importantly, this technique can be applied to other diseases as well, allowing for analysis of cohorts that are less extensively characterized or those with low intrinsic power secondary to low sample size.
Learning points
Cluster analysis, which categorizes subjects into groups of “maximal similarity”, serves as a valuable statistical tool for characterizing phenotypic variability in PD cohorts and for correlating phenotypes with specific biomarkers.
We successfully leveraged cluster analysis of PD subjects from one of the largest observational studies of people with PD, the PPMI study, in order to examine the generalizability of cluster analysis results obtained using a smaller research cohort (Discovery cohort).
The phenotypes derived from clustering analysis of the PPMI cohort included: 1) severe and predominantly nonmotor symptoms with trend towards PIGD phenotype; 2) younger age of onset with overall milder symptoms (both motor and nonmotor); and 3) older age of onset with predominantly motor symptoms and trend towards TD phenotype.
Our results suggest that cluster analysis of large, well-characterized cohorts can be used to establish reproducibility in smaller cohorts with low intrinsic power.
| Supplementary Material | ▴Top |
Suppl 1. Infographic outlining the study methods.
Suppl 2. Barplot depicting missingness in the Discovery cohort, with y-axis representing percent of subjects that did not have data available for a given variable.
Suppl 3. Summary of factor loadings of the first four principal components of the Discovery cohort’s principal component analysis.
Suppl 4. Barplot depicting missingness in the PPMI cohort, with y-axis representing percent of subjects that did not have data available for a given variable.
Suppl 5. Scree plot depicting eigenvectors of PPMI cohort principal components (PC), with an arrow indicating inflection point (elbow) at PC4.
Suppl 6. Summary of factor loadings of the first four principal components of the PPMI cohort’s principal component analysis.
Suppl 7. Principal component analysis of the Discovery cohort (n = 179) was conducted based on traits determined to contribute most significantly to the PPMI cohort. (A) Scatter plot depicting separation of subjects across the first two principal components. (B) Heatmap of clinical characteristics for each cluster identified. The greater red hue indicates greater prevalence, greater blue hue indicates lower prevalence.
Suppl 8. Comparison of demographic and clinical characteristics of the two clusters from the Discovery cohort after repeated cluster analysis using only the traits that drove clustering in the PPMI cohort.
Acknowledgments
We thank all the patients at NYU and MSMD who took part in this study and the participants who took part in the PPMI study, as well as the PPMI database for access to the shared data, and the Nichols Research Lab at the Cincinnati Children’s Hospital for assisting with genotype analyses.
Financial Disclosure
G.R. is supported by grants from the National Institute of Health (NIH) (R01NS133742-01), Michael J Fox Foundation, Parkinson’s Foundation, and Department of Defense (PD210038). T.R. and G.R. are supported by a grant from the NIH (R01NS116006); K.W. is supported by a grant from the Icahn School of Medicine Summer Student Investigator award. J.F.C. is supported by NIH grants RF1NS095252, RF1AG060961, R01NS086736, R01AG062348, R01AG054008, P30AG066514, and U54NS115266, as well as the Rainwater Charitable Foundation (Tau Consortium), and the Parkinson’s Disease Foundation. PPMI (a public-private partnership) is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including Abbvie, Acurex Therapeutics, Allergan, Amathus therapeutics, Avid radiopharmaceuticals, Bial Biotech, Biogen, Biolegend, Bristol Myers Squibb, Calico, Celgene, Jenali, 4D Pharma PLC, GE Healthcare, Genentech, Glass Smith Kline, Golub Capital, Handl Therapeutics, Insitro, Jannsen Neuroscience, Lilli,Lundbeck, Merck, Meso Scale Discovery, Neurocrine Biosciences, Pfizer, Piramal, Prevail Therapeutics, Roche, Sanofi Genzyme, Servier, Takeda, Teva Pharmaceuticals, Union Chimique Belge, Verily, and Voyager Therapeutics.
Conflict of Interest
The authors have no conflict of interest to disclose.
Informed Consent
All the procedures involving human subjects were performed upon written informed consent, approval from the Institutional Review Board and in accord with the Helsinki Declaration of 1975.
Author Contributions
Kristen Watkins: study design, data analysis, manuscript writing and revision. Julia Greenberg: study design, data interpretation, manuscript writing and revision. Kelly Astudillo: data collection, manuscript revision. Charalambos Argyrou: data analysis, manuscript revision. Wen-Yu Lee: data analysis. John F. Crary: data analysis, manuscript revision. Steven J. Frucht: study design, manuscript revision. Towfique Raj: study design, data analysis, manuscript revision. Giulietta Maria Riboldi: study design, data analysis and interpretation, manuscript writing and revision.
Data Availability
The data supporting the findings are available from the corresponding author upon reasonable request. Data used in the preparation of this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org.
Abbreviations
PD: Parkinson’s disease; PPMI: Parkinson’s Progression Markers Initiative; MSMD: Mount Sinai Movement Disorders; NYU: New York University; FoG: freezing of gait; RBD: REM-sleep behavior disorder; MAOi: monoamine oxidase inhibitors; COMT: catechol-O-methyltransferase; TD: tremor dominant; PIGD: postural instability and gait disorder; GBA: glucocerebrosidase; LRRK2: leucine-rich repeat kinase 2; H&Y: Hoehn and Yahr scale; UPSIT: University of Pennsylvania Smell Identification Test; UPDRS III: Unified Parkinson’s Disease Rating Scale Part III; MoCA: Montreal Cognitive Assessment; MDS-UPDRS: Movement Disorder Society - Unified Parkinson’s Disease Rating Scale; GDS: Geriatric Depression Scale; STAI: State-Trait Anxiety Inventory; SCOPA-AUT: Scales for Outcome in Parkinson’s Disease - Autonomic; ESS: Epworth Sleepiness Scale; QUIP: Questionnaire for Impulsive-Compulsive Disorders in Parkinson’s Disease; PCA: principal component analysis; PC: principal component; ANOVA: analysis of variance; REM: rapid eye movement
| References | ▴Top |
- Fereshtehnejad SM, Zeighami Y, Dagher A, Postuma RB. Clinical criteria for subtyping Parkinson's disease: biomarkers and longitudinal progression. Brain. 2017;140(7):1959-1976.
doi pubmed - van Rooden SM, Heiser WJ, Kok JN, Verbaan D, van Hilten JJ, Marinus J. The identification of Parkinson's disease subtypes using cluster analysis: a systematic review. Mov Disord. 2010;25(8):969-978.
doi pubmed - Hendricks RM, Khasawneh MT. A systematic review of Parkinson's disease cluster analysis research. Aging Dis. 2021;12(7):1567-1586.
doi pubmed pmc - Mestre TA, Eberly S, Tanner C, Grimes D, Lang AE, Oakes D, Marras C. Reproducibility of data-driven Parkinson's disease subtypes for clinical research. Parkinsonism Relat Disord. 2018;56:102-106.
doi pubmed - Erro R, Picillo M, Vitale C, Palladino R, Amboni M, Moccia M, Pellecchia MT, et al. Clinical clusters and dopaminergic dysfunction in de-novo Parkinson disease. Parkinsonism Relat Disord. 2016;28:137-140.
doi pubmed - Goetz CG, Poewe W, Rascol O, Sampaio C, Stebbins GT, Counsell C, Giladi N, et al. Movement Disorder Society Task Force report on the Hoehn and Yahr staging scale: status and recommendations. Mov Disord. 2004;19(9):1020-1028.
doi pubmed - Mehanna R, Jankovic J. Young-onset Parkinson's disease: Its unique features and their impact on quality of life. Parkinsonism Relat Disord. 2019;65:39-48.
doi pubmed - van Buuren S, Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations in R. Journal of Statistical Software. 2011;45(3):1-67.
doi - Kowarik A, Templ M. Imputation with the R package VIM. Journal of Statistical Software. 2016;74(7):1-16.
doi - R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2019. https://www.R-project.org/.
- Parkinson Progression Marker Initiative. The parkinson progression marker initiative (PPMI). Prog Neurobiol. 2011;95(4):629-635.
doi pubmed pmc - Zeighami Y, Fereshtehnejad SM, Dadar M, Collins DL, Postuma RB, Dagher A. Assessment of a prognostic MRI biomarker in early de novo Parkinson's disease. Neuroimage Clin. 2019;24:101986.
doi pubmed pmc - Liu R, Umbach DM, Troster AI, Huang X, Chen H. Non-motor symptoms and striatal dopamine transporter binding in early Parkinson's disease. Parkinsonism Relat Disord. 2020;72:23-30.
doi pubmed pmc - Stebbins GT, Goetz CG, Burn DJ, Jankovic J, Khoo TK, Tilley BC. How to identify tremor dominant and postural instability/gait difficulty groups with the movement disorder society unified Parkinson's disease rating scale: comparison with the unified Parkinson's disease rating scale. Mov Disord. 2013;28(5):668-670.
doi pubmed - Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: an R package for determining the relevant number of clusters in a data set. Journal of Statistical Software. 2014;61(6):1-36.
doi - Funtikova AN, Benitez-Arciniega AA, Fito M, Schroder H. Modest validity and fair reproducibility of dietary patterns derived by cluster analysis. Nutr Res. 2015;35(3):265-268.
doi pubmed - McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012;22(3):276-282.
pubmed pmc - McShane LM, Radmacher MD, Freidlin B, Yu R, Li MC, Simon R. Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics. 2002;18(11):1462-1469.
doi pubmed - Kapp AV, Tibshirani R. Are clusters found in one dataset present in another dataset? Biostatistics. 2007;8(1):9-31.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Neurology Research is published by Elmer Press Inc.
